Tracking cancer evolution from cell-free DNA samples
Cell-free DNA (cfDNA) samples offer an unprecedented opportunity to gain a glimpse of tumour composition and evolution without invasive biopsies, which enables frequent sampling in order to real-time follow cancer evolution. However, the inherent heterogeneity and sometimes sub-optimal quality of liquid biopsies mean tracking single nucleotide variants is often not feasible due to high sequencing costs and contamination from healthy DNA. Copy number alterations (CNAs) on the other hand are typically exclusive to tumour cells and can be reliably detected using low-cost shallow whole-genome sequencing even in low quality (e.g. archival) or low tumour content (e.g. cfDNA) materials.
In recent work, I developed a computational method, called liquidCNA, to track subclonal evolution from longitudinally collected cfDNA samples using CNAs. Using liquidCNA, we could show that liquid biopsies and CNAs can track cancer evolution and progression in ovarian cancer.
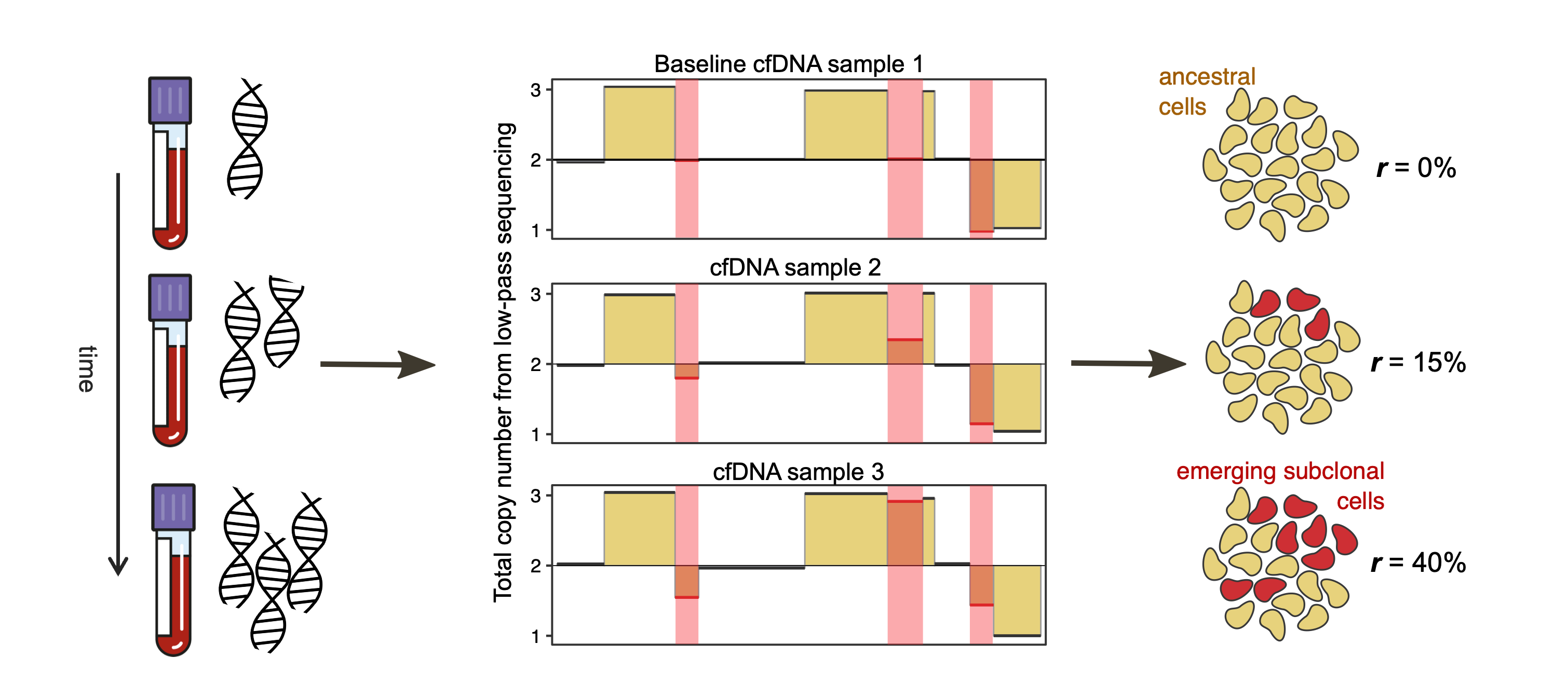
We are interested in improving on this method to make quantative predictions available for a wider set of samples. To this aim, our current projects include
(i) improving tumour DNA detection within cfDNA samples
(ii) improving CNA detection and quantification from low-tumour content samples
(iii) employing mechanistic models to distinguish between evolutionarily relevant signal and measurement noise