The immuno-genomic landscape of cancers
During tumour development, cancer cells continuously acquire mutations that can also give rise to neoantigens, novel cancer-specific peptides presented on the cell surface eliciting a host immune response. Cells that display strongly immunogenic neoantigens are more likely to be eliminated by the immune system, hence neoantigens are associated with a negative selection pressure. In many cancers, however, tumour-associated immune activity is inhibited by genetic alterations in (neo)antigen presentation or modulation of the tumour microenvironment.
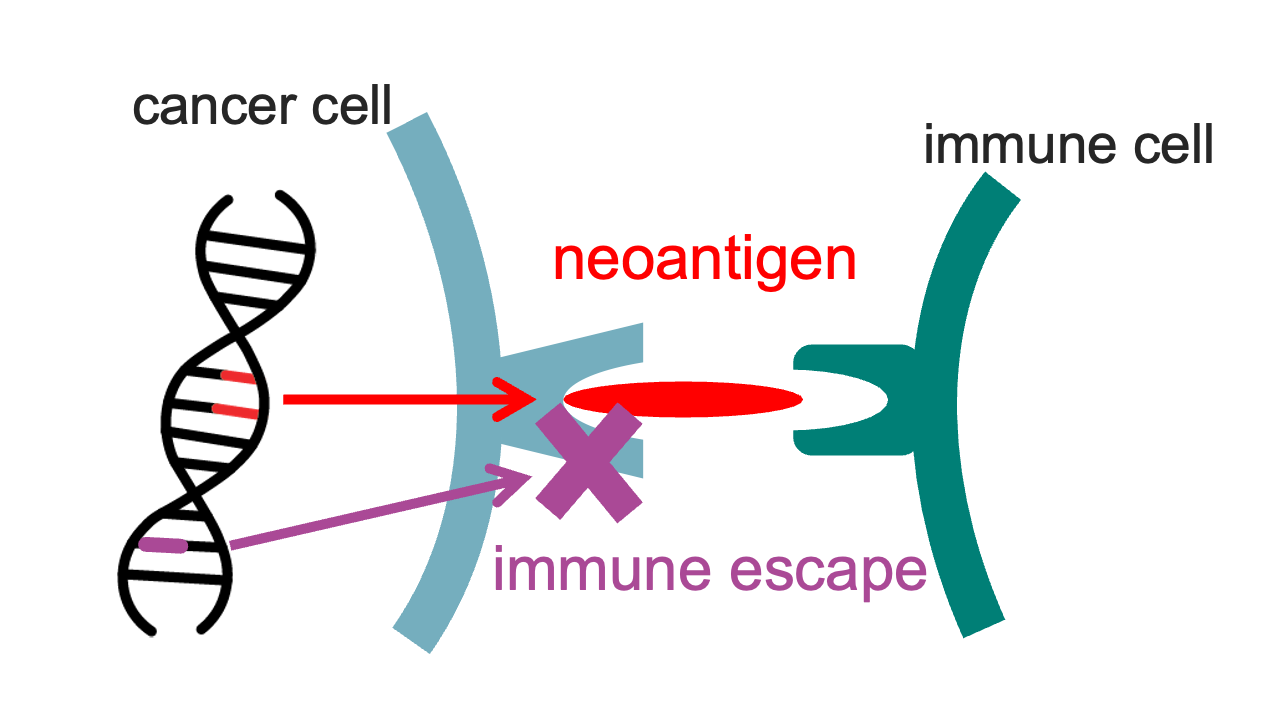
We combine mathematical modelling and bioinformatic evaluation of neoantigens and immune escape to investigate the genetic and epigenetic changes introduced by the tumour-immune interaction. In previous work, I
(i) co-developed a python-based neoantigen prediction pipeline, NeoPredPipe
(ii) identified the pattern of immune-induced negative selection in the clone size (or variant allele frequency) distribution of cancers
(iii) quantified the effect of immune escape mutations in colorectal cancers
(iv) demonstrated the evolutionary advantage arising from mutation rate evolution in mismatch repair-deficient cancers
I lead the Immune Analysis working group of the colorectal, endometrial and testicular cancer domain within the 100,000 Genomes Project.
At the moment, I am focusing on uncovering the epigenetic regulation of cancer immunogenicity through analysing our spatially resolved multi-omic dataset from the Evolutionary Predictions In Colorectal Cancer project. We are interested in uncovering how the ongoing war with the immune system shapes the mutation landscape of colorectal cancers, and if there are distinguished times or locations where immune selection has a stronger impact.
Our other interests include
(i) uncovering the impact of therapy on the cancer-immune interaction
(ii) modelling and quantifying the impact of genetic and epigenetic escape mechanisms
(iii) improving computational predictions of immunogenicity